Investigating the Druggability of SARS-CoV-2 Nucleocapsid Protein RNA interactions

An ETH Zurich research group of the NRP 78 investigated the druggability of the SARS-CoV-2 Nucleocapsid protein RNA interactions. They identified primary fragment hits. Their findings were published in Nucleic Acids Research.
Besides effective vaccines, antiviral compounds represent a fundamental approach to combat viral diseases. The NRP 78 research group of Frédéric Allain, Alvar Gossert and Alexander Leitner at the ETH Zurich investigated whether the interactions between the SARS-CoV-2 nucleocapsid protein, which possesses two known RNA binding domains, and the sm2 RNA located in the 3'UTR of the SARS-CoV-2 RNA could be druggable.
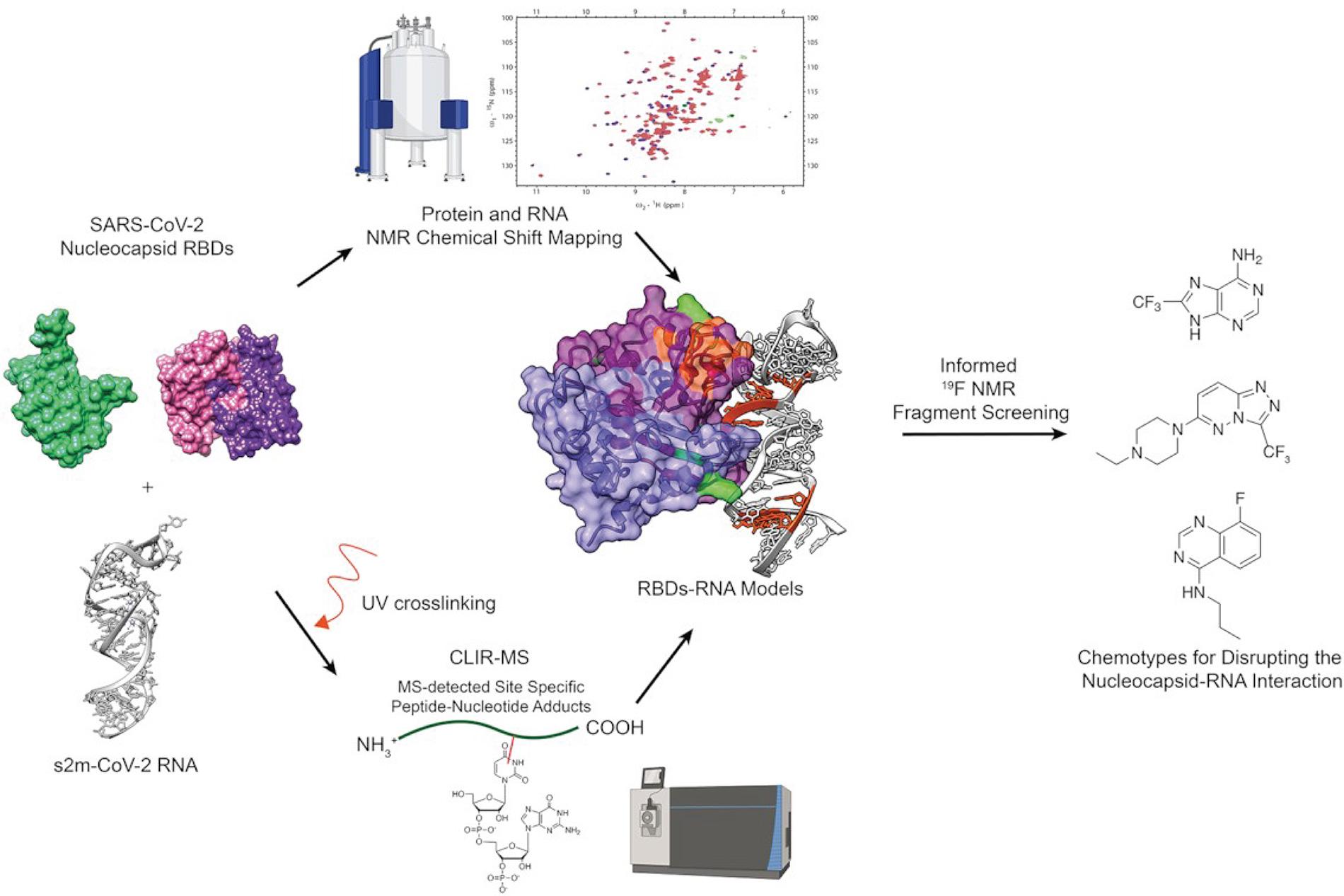
They opted for a hybrid structural approach based on solution NMR experiments and crosslinking MS to obtain structural information on these complexes rapidly. This information was then used to generate restraints to build models of the complexes of the sm2 RNA and the two RNA binding domains of the nucleocapsid protein, for which the unbound structures were solved previously. This approach resulted in the first model of how one of the RNA binding domains interacts with RNA and expanded the knowledge on the other RNA binding domain.
Then an NMR-based fragments screen was used to identify compounds binding to either one of the two protein domains or the RNA in isolation. From these compounds, the ones were selected that bind to the interaction interface of the respective molecules, which were known from the hybrid structural approach models. Overall, the experiments revealed that these protein RNA interactions seem druggable. Therefore, the identified compounds provide the basis to develop more potent molecules inhibiting these protein-RNA interactions.
